Perform principal component analysis using assays or the joint probability matrix as input.
Usage
RunPCA.SingleCellExperiment(
object,
assay.name,
p,
scale,
center,
threshold,
pca.method,
return.model,
select.icp.tables,
features,
dimred.name
)
# S4 method for class 'SingleCellExperiment'
RunPCA(
object,
assay.name = "joint.probability",
p = 50,
scale = TRUE,
center = TRUE,
threshold = 0,
pca.method = "irlba",
return.model = FALSE,
select.icp.tables = NULL,
features = NULL,
dimred.name = "PCA"
)Arguments
- object
A
SingleCellExperimentobject.- assay.name
Name of the assay to compute PCA. One of
assayNames(object)orjoint.probability. By defaultjoint.probabilityis used. Usejoint.probabilityto obtain an integrated embedding after runningRunParallelDivisiveICP. One of the assays inassayNames(object)can be provided before performing integration to assess if data requires integration.- p
A positive integer denoting the number of principal components to calculate and select. Default is
50.- scale
A logical specifying whether the probabilities should be standardized to unit-variance before running PCA. Default is
TRUE.- center
A logical specifying whether the probabilities should be centered before running PCA. Default is
TRUE.- threshold
A threshold for filtering out ICP runs before PCA with the lower terminal projection accuracy below the threshold. Default is
0.- pca.method
A character specifying the PCA method. One of
"irlba"(default),"RSpectra"or"stats". Set seed before, if the method is"irlba"to ensure reproducibility.- return.model
A logical specifying if the PCA model should or not be retrieved. By default
FALSE. Only implemented forpca.method = "stats". IfTRUE, thepca.methodis coerced to"stats".- select.icp.tables
Select the ICP cluster probability tables to perform PCA. By default
NULL, i.e., all are used, except if the ICP tables were obtained with the functionRunParallelDivisiveICP, in which the ICP tables correspond to the last round of divisive clustering for every epoch. A vector ofintegersshould be given otherwise.- features
A character of feature names matching
row.names(object)to select from before computing PCA. Only used ifassay.nameis one of the assays inassayNames(object), otherwise it is ignored.- dimred.name
Dimensional reduction name given to the returned PCA. By default
"PCA".
Examples
# Import package
suppressPackageStartupMessages(library("SingleCellExperiment"))
# Create toy SCE data
batches <- c("b1", "b2")
set.seed(239)
batch <- sample(x = batches, size = nrow(iris), replace = TRUE)
sce <- SingleCellExperiment(assays = list(logcounts = t(iris[,1:4])),
colData = DataFrame("Species" = iris$Species,
"Batch" = batch))
colnames(sce) <- paste0("samp", 1:ncol(sce))
# Prepare SCE object for analysis
sce <- PrepareData(sce)
#> Converting object of `matrix` class into `dgCMatrix`. Please note that Coralysis has been designed to work with sparse data, i.e. data with a high proportion of zero values! Dense data will likely increase run time and memory usage drastically!
#> 4/4 features remain after filtering features with only zero values.
# Multi-level integration (just for highlighting purposes; use default parameters)
set.seed(123)
sce <- RunParallelDivisiveICP(object = sce, batch.label = "Batch",
k = 2, L = 25, C = 1, train.k.nn = 10,
train.k.nn.prop = NULL, use.cluster.seed = FALSE,
build.train.set = FALSE, ari.cutoff = 0.1,
threads = 2)
#>
#> Initializing divisive ICP clustering...
#>
|
| | 0%
|
|=== | 4%
|
|====== | 8%
|
|========= | 12%
|
|============ | 17%
|
|=============== | 21%
|
|================== | 25%
|
|==================== | 29%
|
|======================= | 33%
|
|========================== | 38%
|
|============================= | 42%
|
|================================ | 46%
|
|=================================== | 50%
|
|====================================== | 54%
|
|========================================= | 58%
|
|============================================ | 62%
|
|=============================================== | 67%
|
|================================================== | 71%
|
|==================================================== | 75%
|
|======================================================= | 79%
|
|========================================================== | 83%
|
|============================================================= | 88%
|
|================================================================ | 92%
|
|=================================================================== | 96%
|
|======================================================================| 100%
#>
#> Divisive ICP clustering completed successfully.
#>
#> Predicting cell cluster probabilities using ICP models...
#> Prediction of cell cluster probabilities completed successfully.
#>
#> Multi-level integration completed successfully.
# Integrated PCA
set.seed(125) # to ensure reproducibility for the default 'irlba' method
sce <- RunPCA(object = sce, assay.name = "joint.probability", p = 10)
#> Divisive ICP: selecting ICP tables multiple of 1
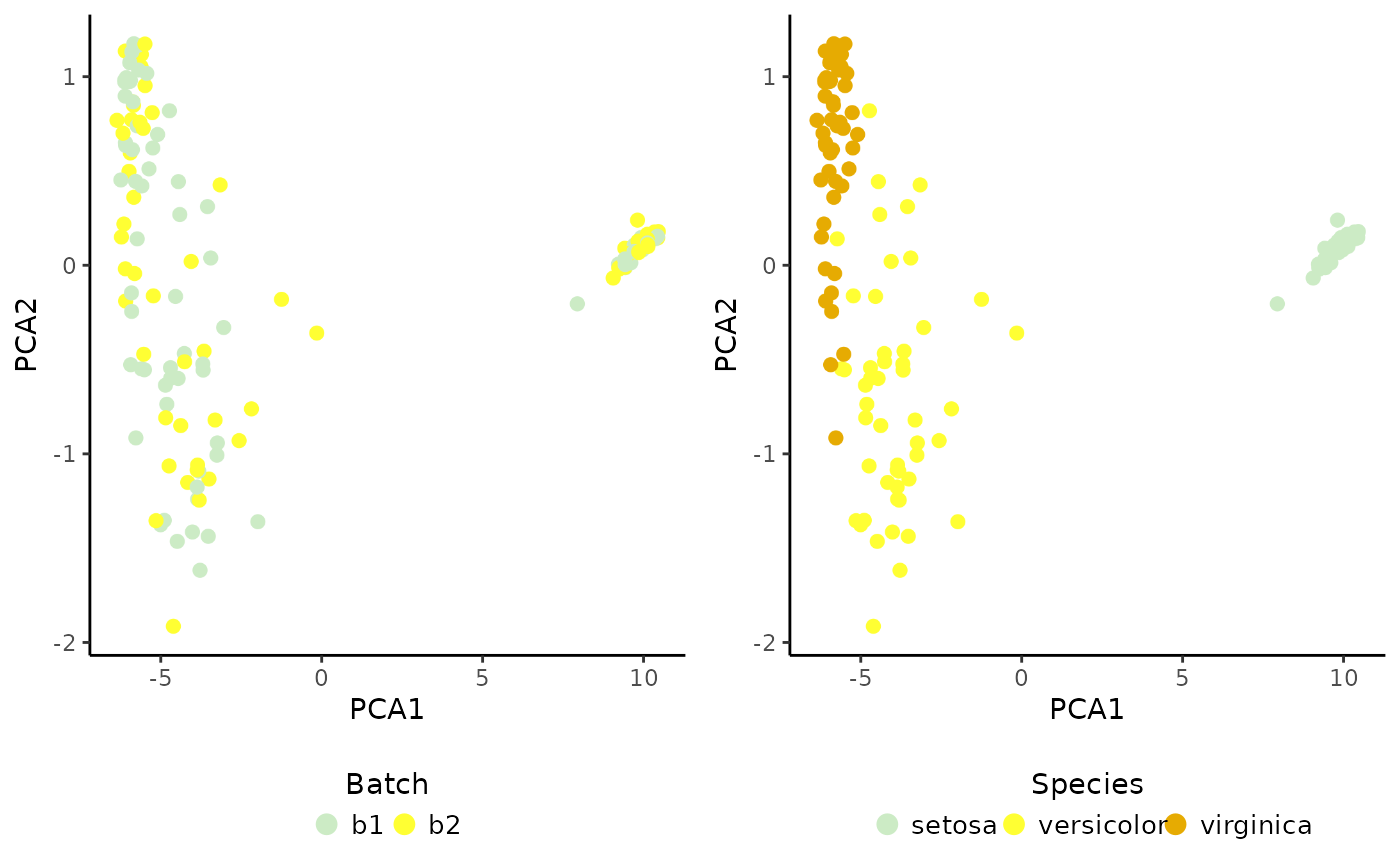
# Plot result
cowplot::plot_grid(PlotDimRed(object = sce, color.by = "Batch",
legend.nrow = 1),
PlotDimRed(object = sce, color.by = "Species",
legend.nrow = 1), ncol = 2)