Protocols
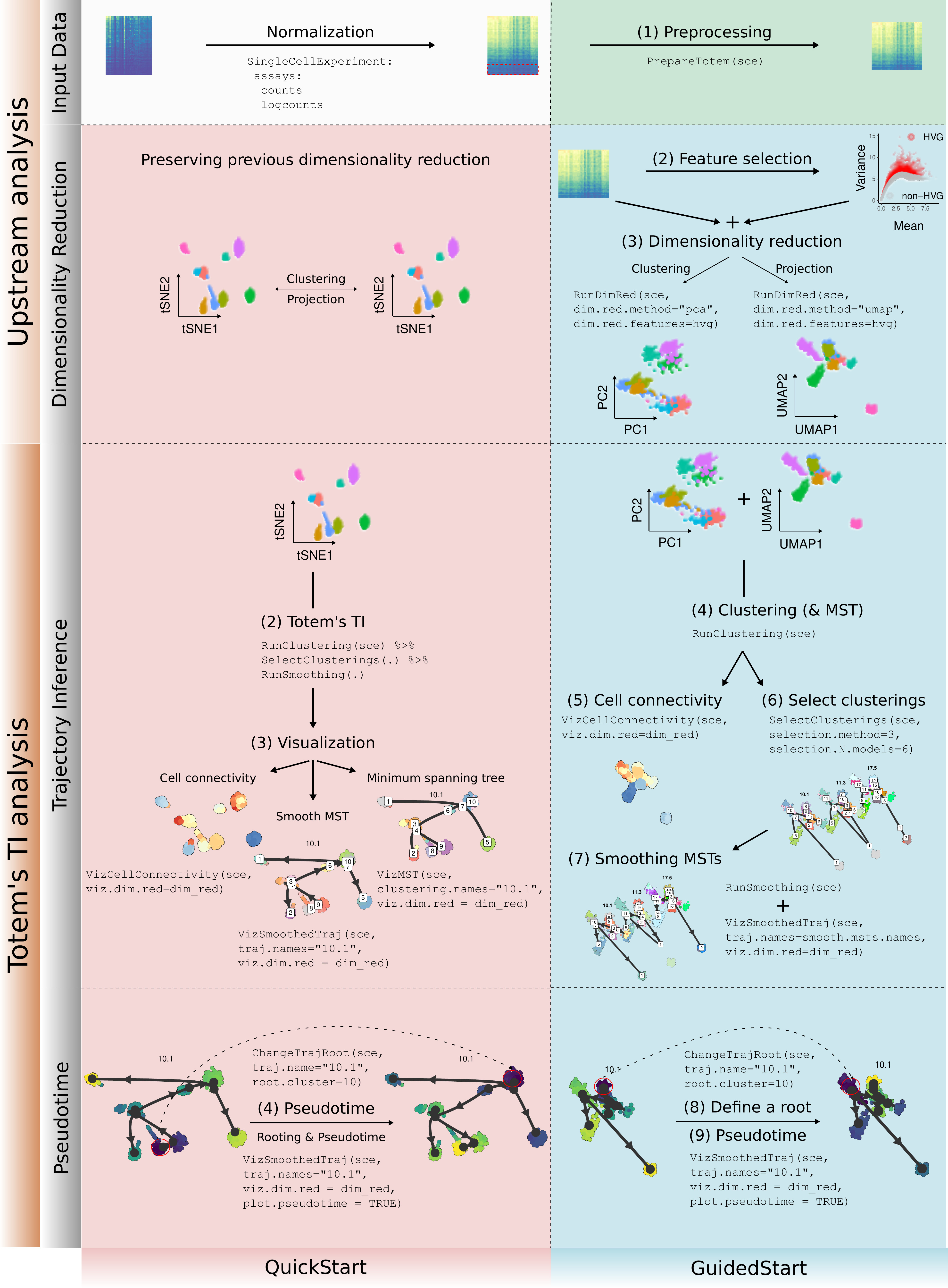
Links for two user-friendly protocols for inferring tree-shaped single-cell trajectories and pseudotime from single-cell transcriptomics data with Totem can be found below: QuickStart and GuidedStart. QuickStart provides a simple and practical example, whereas the GuidedStart protocol details the analysis step-by-step.
All the analyses can be fully reproduced in Linux, MacOS, and Windows OS (amd64 architecture) with >8Gb of RAM using the provided docker image distributed with notebooks, scripts, and data in Docker Hub (elolab/repro-totem-ti).
See the schematic figure below which describes and compares the main step protocols.